We aimed to synthesise our DNA Origami structure and validate the NanoFilter mechanism of action in a medical context.
We firstly synthesised and imaged the core structure of the NanoFilter as it was designed (see NanoFilter Version 1 Synthesis). We next optimised its rigidity to confirm our structure was capable of rolling in the desired fashion (see Optimising NanoFilter Rigidity). After rigidity was optimised, we folded and imaged the structure with locks, such that it curled (see NanoFilter Version 2 Synthesis). We finally determined DNA Origami was sufficiently durable to avoid degradation in liquid flow conditions that would mimic its use in a medical context (see Durability of DNA Origami in Liquid Flow).
Our first experimental goals were to fold a prototype of the NanoFilter, optimise its folding conditions and image it using Transmission Electron Microscopy (TEM). This version did not have handles or locks, so could not curl or capture products. Our aim was to prove that the core staples can self-assemble into the desired structure with seven nodes and six linkers. See here for more experimental details.
We aimed to determine the optimum temperature, salt concentration and linker type to efficiently fold the NanoFilter.
Two types of links between nodes were tested- single stranded links and double stranded links. Single stranded links had no specific linker strands, using the scaffold as the sole connection between nodes. Double stranded links used linker strands complementary to the scaffold to form double stranded DNA between the nodes (see Diagram 1).

We used two different ramps controlling folding temperature- 50-40°C and 65-25°C. These involved reducing the temperature from the highest to lowest in the ramp incrementally over 18 hours. We also tested six concentrations of Magnesium Chloride in our folding buffer- 4, 8, 12, 16, 20 and 24 mM.
All combinations of the conditions were used, and run on an agarose gel.

There were bands that corresponded to the folded DNA origami in all lanes. This indicates all conditions could facilitate NanoFilter folding to a degree.
DNA folded with a 65-25°C ramp was more prone to aggregation than DNA folded with the 50-40°C ramp. This is evident in the presence of faint bands above the brightest DNA Origami band. The clearest and brightest bands were seen in the 16mM salt concentration, although there was not substantial difference between the results from different salt concentrations. Bands were of similar brightness between the two linker types, with the ss link structure migrating further, since it was a smaller structure.
We concluded optimal folding conditions for efficient production of the NanoFilter was when DNA was folded in 16mM of Magnesium Chloride for 18 hours from 50-40°C. Single stranded and double stranded link structures folded with the same efficiency.
We imaged the NanoFilter after folding with its optimal conditions- a 50-40°C ramp and at 16 mM. We initially imaged the ds Link structure (see Figure 2) to confirm the more complex link structure was viable.

The staples and scaffold self-assembled into seven nodes and six links between them. The structure was sufficiently flexible to curl.
We concluded that the structure was as expected, and had sufficient flexibility to be viable in future experiments.
We next investigated which folding conditions allowed for the optimal flexibility of the NanoFilter. This was important, as if the structure is too rigid, it will not be able to roll upon contact with LDL cholesterol. However, if it is too flexible, it may not adopt the desired structure. We investigated whether the linker strand type affected the flexibility of the final structure. See here for experimental details.
Samples from the gel above folded for 18 hours at 50-40°C and in 16 mM Magnesium Chloride were imaged (see Figure 3). The image of the single strand link NanoFilter was less focused than the double strand link NanoFilter.

From all the images taken, it appeared as though the NanoFilter with double-stranded links was sufficienctly flexible to curve, but remained rigid and did not self-aggregate. However, the nanoFilter with single strand linkers appeared to often self-aggregate, and so was likely insufficiently rigid to maintain a fixed structure.
We measured the length of the individual DNA Origami structures with both linker types from a number of TEM images. We took the maximum straight line length present in the Origami (see Diagram 2). In this way, if the NanoFilter was clumped, we would record a smaller in size and if it was stretched out, we would record a longer size.

This was converted into lengths in nanometres using the image scales and graphed (see Figure 4).
An unpaired t test was conducted and found that the structures with ds links were significantly longer than those with ss links (p < 0.001).
We concluded that the double strand link structure could bend, but was more rigid than the single strand link structure and did not self-aggregate as easily. It is is therefore more apt to use in our future experiments.
The second version of the NanoFilter was designed with a locking mechanism that enables it to fold into its spiral form. It is also designed with a catalyst that allows preferential folding in a particular direction. This version was built with staples that were further categorised into three types:
Plain: This structure does not have any handles (so will not be able to close)
Non-Specific: The handles are attached to all nodes and linkers (testing the locking mechanism)
Specific: The handles are present on specific nodes and linkers (testing specific configurations of the spiral)
See here for experimental details.
We folded three samples- NanoFilters with non-specific and specific handles along with their corresponding locks, and a NanoFilter sample with plain handles and no locks. They were folded for 2 hours from 50 to 40°C. Samples were subject to TEM imaging, but unlike for the version 1 NanoFilter, they were not gel purified beforehand.
The staining for NanoFilters with plain or non-specific handles was too strong, and so no image could be obtained. Images of the NanoFilter with handles targetted to specific nodes were taken at a number of magnifications (see Figure 5).

TEM imaging found circular structures assembled which were structurally similar to the designed locked NanoFilter. However, the images were not clear and so difficult to interpret.
We concluded that we had likely folded a locked version of our NanoFilter as per our caDNAno design. In the future we should image purified NanoFilters that were folded over an 18 hour period, purified and had less staining to obtain clearer TEM images.
We conducted experiments to investigate whether DNA Origami degrades when subject to liquid flow. If DNA Origami is not sufficiently durable, then the NanoFilter would degrade once connected to blood vessels, and it would not be useful in a medical context. We used barrels created by the 2018 USyd BIOMOD team as a surrogate in our experiments. See here for details of its structure and function. It was a useful surrogate as we could test the strength of the base pairing in DNA Origami in a system which had been proven to bind glass slides. See here for experimental details.
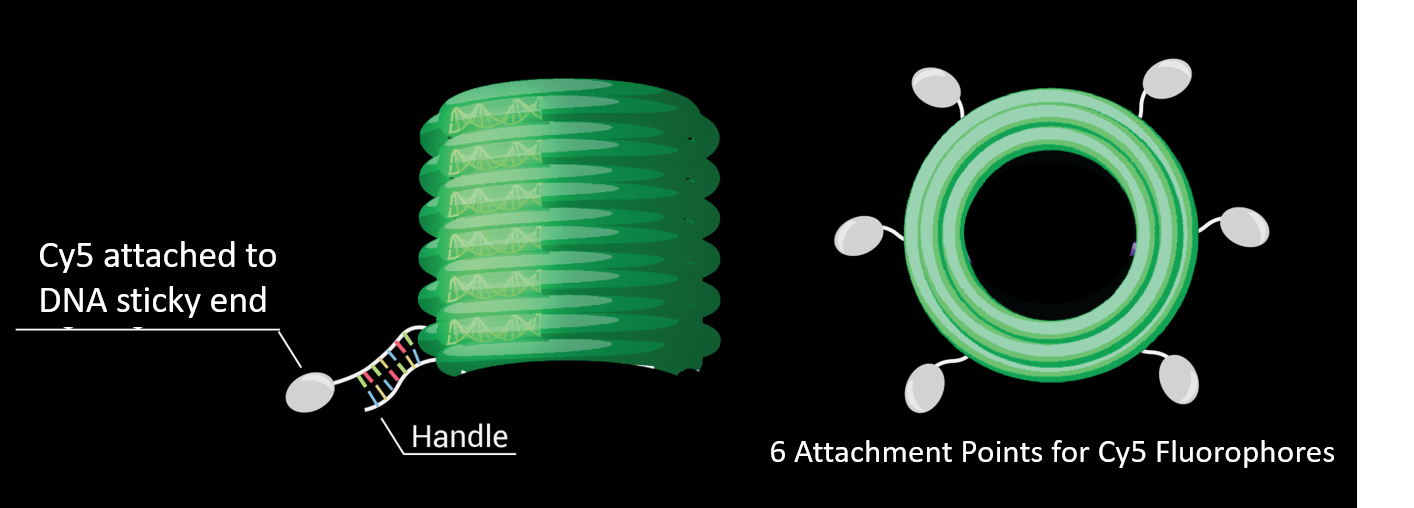
We generated DNA Origami barrels with biotinylated side strands and sticky ends protruding from the outer staples. DNA complementary to the sticky ends and conjugated to Cy5 fluorophores was added to generate fluorescent barrels (see Diagram 3).
We made a flow chamber by putting four layers of water-resistant double-sided tape 4 mm apart on a slide, and placing a cover slip on top. The chamber was incubated with BSA-biotin, Streptavidin then DNA Origami samples with Cy5 Fluorophores (see Diagram 4). The barrels that did not bind were removed by pipette suction and stored.

Magnesium Chloride Wash buffer was flowed into the chamber, removed on the other side and pushed back (see Diagram 5). This was repeated two times, so that the same liquid moved back and forth three times through the chamber. The liquid was stored as 3x flow. This was repeated, but with ten flows and 20 flows.

The samples were run in an agarose gel with SYBR Safe staining. It was imaged at emission wavelengths for Cy5 Fluorescence and Sybr Safe (see Figure 6).

There was a band in the lane loaded with sample of barrels that did not bind, which indicates not all of our sample bound to the glass slide. However, the sample band was much lighter than the no streptavidin control band, which indicates that some barrels bound to the slide. There was no DNA detected in any of the flow samples.
We concluded the DNA origami was durable in flow conditions, as no DNA degraded after even the most significant flow.